
Page 1 of 10
Article DOI: https://doi.org/10.3201/eid2512.190267
Genomic Analysis of Fluoroquinolone- and
Tetracycline-Resistant Campylobacter jejuni
Sequence Type 6964 in Humans and
Poultry, New Zealand, 2014–2016
Appendix
Further details of participating human diagnostic laboratories and poultry farm
survey
The laboratories that referred isolates for the human case survey were Labtests,
Auckland; Pathlab Bay of Plenty; Aotea Pathology, Wellington; Canterbury Southern
Community Laboratories, Christchurch; and Southern Community Laboratories, Dunedin.
Details of the survey are provided are provided in the report by Williamson, Dyet (1).
For the poultry cecal surveys pooled cecal samples (each containing cecal contents from
5 chickens) were taken from 4 poultry companies at slaughter. One study was conducted
between May 25, 2015 and June 02, 2015 and the other between July 07, 2015 and March 25,
2016. A total of 897 pooled cecal samples from broiler and breeder flocks were processed, of
which 53.7% (482/897) were positive for growth typical of Campylobacter on selective media
containing ciprofloxacin and tetracycline. Only six composite samples were received from the
small Company D and none of these samples yielded growth on the mCCDA + cip/tet plates. A
random subset of 291 isolates were speciated by PCR and all were confirmed as C. jejuni. A
further subset of 99 of the 291 isolates were subtyped by 7-gene MLST and all were confirmed
as ST-6964. Given the high probability that isolates cultured on the selective media were C.
jejuni ST-6964, 136 of the 291 PCR confirmed isolates (from 118 pooled samples) were
randomly selected for WGS, and all were confirmed by WGS as ST-6964 (39 by 7-gene MLST
and WGS and 97 by WGS only).

Page 2 of 10
A map showing the location of the participating laboratories and the poultry farms from
which C. jejuni ST 6964 positive cecal samples were taken for analysis is shown in Appendix
Figure 1.
References
1. Williamson D, Dyet K, Heffernan H. Antimicrobial resistance in human isolates of Campylobacter
jejuni, 2015: Institute of Environmental Science and Research Limited; 2015.
2. Marasini D, Fakhr MK. Whole-genome sequencing of a Campylobacter jejuni strain isolated from
retail chicken meat reveals the presence of a megaplasmid with Mu-like prophage and multidrug
resistance genes. Genome Announc. 2016;4:e00460-16. PubMed
https://doi.org/10.1128/genomeA.00460-16
3. Marasini D, Fakhr MK. Complete genome sequences of plasmid-bearing multidrug-resistant
Campylobacter jejuni and Campylobacter coli strains with type VI secretion systems, isolated
from retail turkey and pork. Genome Announc. 2017;5:e01360-17. PubMed
https://doi.org/10.1128/genomeA.01360-17
4. Marasini D, Fakhr MK. Complete genome sequences of Campylobacter jejuni strains isolated from
retail chicken and chicken gizzards. Genome Announc. 2017;5:e01351-17. PubMed
https://doi.org/10.1128/genomeA.01351-17
5. Crespo MD, Altermann E, Olson J, Miller WG, Chandrashekhar K, Kathariou S. Novel plasmid
conferring kanamycin and tetracycline resistance in the turkey-derived Campylobacter jejuni
strain 11601MD. Plasmid. 2016;86:32–7. PubMed https://doi.org/10.1016/j.plasmid.2016.06.001
6. Gunther NW IV, Reichenberger ER. Complete genome sequence of Campylobacter jejuni RM1246-
ERRC, which exhibits resistance to quaternary ammonium compounds. Genome Announc.
2017;5:e00978-17. PubMed https://doi.org/10.1128/genomeA.00978-17
7. Batchelor RA, Pearson BM, Friis LM, Guerry P, Wells JM. Nucleotide sequences and comparison of
two large conjugative plasmids from different Campylobacter species. Microbiology.
2004;150:3507–17. PubMed https://doi.org/10.1099/mic.0.27112-0
8. Marasini D, Fakhr MK. Complete genome sequences of Campylobacter jejuni strains OD267 and
WP2202 isolated from retail chicken livers and gizzards reveal the presence of novel 116-
Kilobase and 119-Kilobase megaplasmids with type VI secretion systems. Genome Announc.
2016;4:e01060-16. PubMed https://doi.org/10.1128/genomeA.01060-16

Page 3 of 10
9. Taveirne ME, Dunham DT, Perault A, Beauchamp JM, Huynh S, Parker CT, et al. Complete annotated
genome sequences of three Campylobacter jejuni strains isolated from naturally colonized farm-
raised chickens. Genome Announc. 2017;5:e01407-16. PubMed
https://doi.org/10.1128/genomeA.01407-16
10. Zhang M, He L, Li Q, Sun H, Gu Y, You Y, et al. Genomic characterization of the Guillain-Barre
syndrome-associated Campylobacter jejuni ICDCCJ07001 Isolate. PLoS One. 2010;5:e15060.
PubMed https://doi.org/10.1371/journal.pone.0015060
11. Cooper KK, Cooper MA, Zuccolo A, Law B, Joens LA. Complete genome sequence of
Campylobacter jejuni strain S3. J Bacteriol. 2011;193:1491–2. PubMed
https://doi.org/10.1128/JB.01475-10
Appendix Table 1. Details of the plasmids used in the comparative analysis of plasmid 15AR0984-m from the reference strain of
ST-6964 used in this study
Accession no.
Reference
Description
CP013117.1
(2)
C. jejuni strain T1–21 plasmid pcjDM
CP017857.1
(3)
C. jejuni strain YQ2210 plasmid pCJDM210L
CP017854.1
(4)
C. jejuni strain ZP3204 plasmid pCJDM204L
KJ646012.1
(5)
C. jejuni strain 11601MD plasmid p11601MD
CP022471.1
(6)
C. jejuni strain RM1246-ERRC plasmid pRM1246_ERRC
AY394561.1
(7)
C. jejuni strain 81–176 plasmid pTet
CP014745.1
(8)
C. jejuni strain OD267 plasmid pCJDM67 L
CP017418.1
(9)
C. jejuni strain MTVDSCj13 plasmid pMTVDSCj13–1
CP007750.1
Not applicable
C. jejuni strain M129 plasmid pTet-M129
CP007182.1
Not applicable
C. coli strain RM4661 plasmid pRM4661
CP002030.1
(10)
C. jejuni strain ICDCCJ07001 plasmid pTet
CP001961.1
(11)
C. jejuni strain S3 plasmid pTet

Page 4 of 10
Appendix Table 2. The 87 wgMLST shared-loci that were within the Gubbins predicted recombination regions. The aliases in the
first column correspond to the gene names used in the wgMLST analysis.
Alias
Gene
Function
Locus tag
Gene577
Hypothetical protein
15AR0984_00592
Gene1430
Peptidylprolyl isomerase
15AR0984_01465
Gene1431
accC
Biotin carboxylase
15AR0984_01466
Gene1432
accB
Biotin carboxyl carrier protein of acetyl-coa carboxylase
15AR0984_01467
Gene1433
dcd
Deoxycytidine triphosphate deaminase
15AR0984_01468
Gene1434
pseB
UDP-N-acetylglucosamine 4,6-dehydratase (inverting)
15AR0984_01469
Gene1435
pseC
UDP-4-amino-4, 6-dideoxy-N-acetyl-β-L-altrosamine transaminase
15AR0984_01470
Gene1437
Putative polysaccharide biosynthesis protein with aminopeptidase-
like domain protein
15AR0984_01472
Gene1441
Hypothetical protein
15AR0984_01476
Gene1442
fabH
3-oxoacyl-[acyl-carrier-protein] synthase 3
15AR0984_01477
Gene1443
Hypothetical protein
15AR0984_01478
Gene1444
Hypothetical protein
15AR0984_01479
Gene1445
Hypothetical protein
15AR0984_01480
Gene1449
Hypothetical protein
15AR0984_01484
Gene1454
legF
CMP-N,N'-diacetyllegionaminic acid synthase
15AR0984_01489
Gene1455
pseG
UDP-2,4-diacetamido-2,4, 6-trideoxy-β-L-altropyranose hydrolase
15AR0984_01490
Gene1456
pseH
UDP-4-amino-4, 6-dideoxy-N-acetyl-β-L-altrosamine N-
acetyltransferase
15AR0984_01491
Gene1457
hisF
Imidazole glycerol phosphate synthase subunit hisf
15AR0984_01492
Gene1458
hisH1
Imidazole glycerol phosphate synthase subunit hish 1
15AR0984_01493
Gene1459
Hypothetical protein
15AR0984_01494
Gene1461
rmlB
Dtdp-glucose 4,6-dehydratase
15AR0984_01496
Gene1464
legI
N,N'-diacetyllegionaminic acid synthase
15AR0984_01499
Gene1469
hddC
D-glycero-α-D-manno-heptose 1-phosphate guanylyltransferase
15AR0984_01504
Gene1470
Hypothetical protein
15AR0984_01505
Gene1471
legF
CMP-N,N'-diacetyllegionaminic acid synthase
15AR0984_01506
Gene1472
lvr
Levodione reductase
15AR0984_01507
Gene1476
Hypothetical protein
15AR0984_01511
Gene1481
Hypothetical protein
15AR0984_01516
Gene1482
Hypothetical protein
15AR0984_01517
Gene1483
xcpT
Type II secretion system protein G
15AR0984_01518
Gene1484
tsaD
Trna N6-adenosine threonylcarbamoyltransferase
15AR0984_01519
Gene1486
dxr
1-deoxy-D-xylulose 5-phosphate reductoisomerase
15AR0984_01521
Gene1487
cdsA
Phosphatidate cytidylyltransferase
15AR0984_01522
Gene1488
Hypothetical protein
15AR0984_01523
Gene1489
Hypothetical protein
15AR0984_01524
Gene1491
Putative phospholipase A1
15AR0984_01526
Gene1492
feuB
Iron-uptake system permease protein feub
15AR0984_01527
Gene1493
feuC
Iron-uptake system permease protein feuc
15AR0984_01528
Gene1494
fhuC
Iron(3+)-hydroxamate import ATP binding protein fhuc
15AR0984_01529
Gene1496
Hypothetical protein
15AR0984_01532
Gene1497
nrfA
Cytochrome c-552
15AR0984_01533
Gene1498
nrfH
Cytochrome c-type protein nrfh
15AR0984_01534
Gene1499
ppk
Polyphosphate kinase
15AR0984_01535
Gene1504
Hypothetical protein
15AR0984_01541
Gene1505
ruvB
Holliday junction ATP-dependent DNA helicase ruvb
15AR0984_01542
Gene1507
fumC
Fumarate hydratase class II
15AR0984_01544
Gene1509
glmS
Glutamine–fructose-6-phosphate aminotransferase [isomerizing]
15AR0984_01546
Gene1511
mqnE
Aminodeoxyfutalosine synthase
15AR0984_01548
Gene1512
adeQ
Adenine permease adeq
15AR0984_01549
Gene1513
pyrE
Orotate phosphoribosyltransferase
15AR0984_01550
Gene1514
mlaA
Putative phospholipid binding lipoprotein mlaa
15AR0984_01551
Gene1515
mlaC
Putative phospholipid binding protein mlac
15AR0984_01552
Gene1516
Bifunctional preprotein translocase subunit secd/secf
15AR0984_01553
Gene1589
kpsM
Polysialic acid transport protein kpsm
15AR0984_01626
Gene1590
Hypothetical protein
15AR0984_01627
Gene1591
Hypothetical protein
15AR0984_01628
Gene1592
Dutpase
15AR0984_01629
Gene1593
Hypothetical protein
15AR0984_01630
Gene1595
rimO
Ribosomal protein S12 methylthiotransferase rimo
15AR0984_01632
Gene1596
prfB
Peptide chain release factor 2
15AR0984_01633
Gene1597
Hypothetical protein
15AR0984_01634
Gene1598
truD
Trna pseudouridine synthase D
15AR0984_01635
Gene1599
thiL
Thiamine-monophosphate kinase
15AR0984_01636
Gene1601
Hypothetical protein
15AR0984_01638

Page 5 of 10
Alias
Gene
Function
Locus tag
Gene1602
rsmD
Ribosomal RNA small subunit methyltransferase D
15AR0984_01639
Gene1603
flgI
Flagellar P-ring protein
15AR0984_01640
Gene1604
Hypothetical protein
15AR0984_01641
Gene1605
Anti-sigma-28 factor, flgm
15AR0984_01642
Gene1606
Flgn protein
15AR0984_01643
Gene1607
flgK
Flagellar hook-associated protein 1
15AR0984_01644
Gene1609
Hypothetical protein
15AR0984_01646
Gene1616
por
Pyruvate synthase
15AR0984_01653
Gene1617
ppaX
Pyrophosphatase ppax
15AR0984_01654
Gene1618
oprF
Outer membrane porin F
15AR0984_01655
Gene1619
rpsI
30S ribosomal protein S9
15AR0984_01656
Gene1620
rplM
50S ribosomal protein L13
15AR0984_01657
Gene1621
addA
ATP-dependent helicase/nuclease subunit A
15AR0984_01658
Gene1624
Hypothetical protein
15AR0984_01661
Gene1625
Hypothetical protein
15AR0984_01662
Gene1626
Hypothetical protein
15AR0984_01663
Gene1627
fixP
Cbb3-type cytochrome c oxidase subunit fixp
15AR0984_01664
Gene1628
Cbb3-type cytochrome oxidase component fixq
15AR0984_01665
Gene1629
Cytochrome C oxidase, mono-heme subunit/fixo
15AR0984_01666
Gene1630
ccoN1
Cbb3-type cytochrome c oxidase subunit ccon1
15AR0984_01667
Gene1631
mprA
Response regulator mpra
15AR0984_01668
Gene1640
Hypothetical protein
15AR0984_01678
Gene1641
putP
Sodium/proline symporter
15AR0984_01679
Appendix Figure 1. The location of the poultry farms from which C. jejuni ST 6964 positive cecal
samples were taken and the diagnostic laboratories that submitted human isolates including the 5 that
participated in the human survey and the MidCentral laboratory in the Manawatu sentinel site.

Page 6 of 10
Appendix Figure 2. Pairwise distances and NeighborNet networks of the 227 C. jejuni ST-6964 isolates
calculated by two independent wgMLST methods. The upper network was generated from the 1,363
shared loci found in the ad hoc wgMLST analysis; and the lower network was generated with the C. jejuni
cgMLST scheme (1,343 loci) on the PubMLST Web site.

Page 7 of 10
Appendix Figure 3. Recombination detected in 227 C. jejuni ST-6964 isolates. Coding regions are
shown on both the forward and reverse strand of the annotated C. jejuni 15AR0984 reference genome.
The tree is the inferred mid-point rooted phylogeny of 227 C. jejuni ST-6964 isolates and reference
15AR0984 genome is shown to the left and the recombination blocks identified by Gubbins are shown to
the right of the tree; red indicates conserved blocks, blue indicates blocks detected in just one
representative. The lower plot (black line) summarizes the number of recombination events across the
entire C. jejuni reference genome.

Page 8 of 10
Appendix Figure 4. NeighborNet phylogenetic relationship of the plasmid identified in the complete
genome of isolate 15AR0984 related to nine other representative 'pTet-like' plasmids, based on the allele
profiles of the 23 shared-loci. The genome structures of the closest plasmid pcjDM (four allele
differences) and the other three plasmids were plotted to demonstrate the consensus regions (backbone)
and hyper-variable regions in the plasmids.
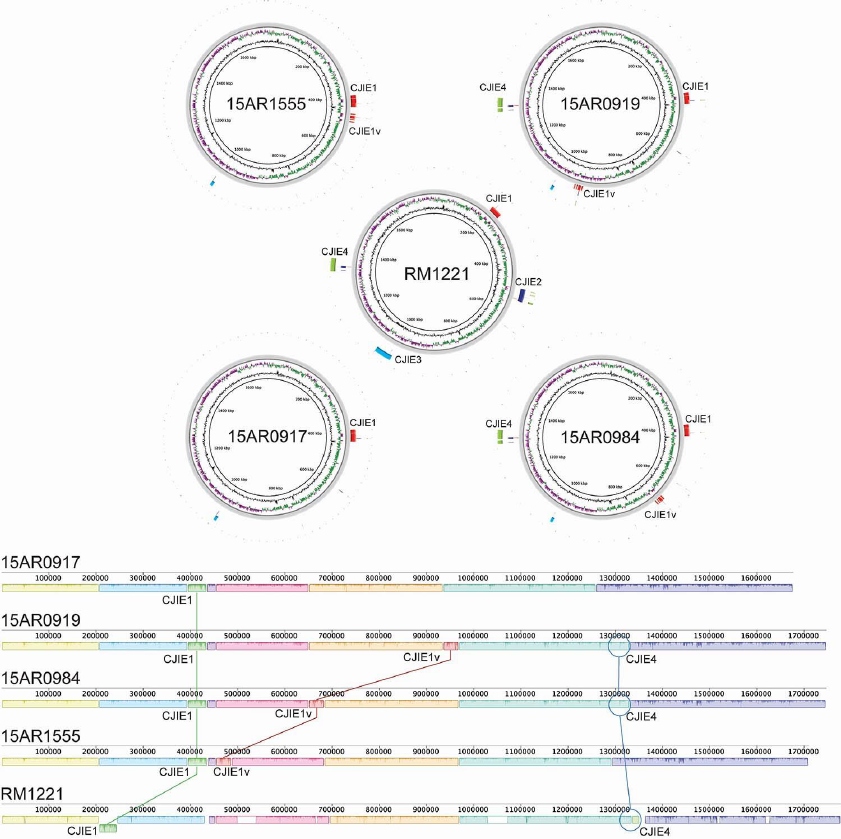
Page 9 of 10
Appendix Figure 5. The insertion locations of mobile elements CJIE1, CJIE1v and CJIE4 are illustrated
in a whole genome alignment of the four complete genomes of ST-6964 isolates (15AR0984, 15AR0917,
15AR0919 and 15AR1555) and the reference RM1221. The upper rings were generated with the BLAST
Ring Image Generator (BRIG) and the lower linear plots with Mauve. The mobile element CJIE1 was
present in all isolates. CJIE1v was inserted in the three different locations in three of the ST-6964 isolate
genomes.

Page 10 of 10
Appendix Figure 6. High-scoring Segment Pairs (HSPs) between the chromosome and plasmid
sequences of the strain 15AR0984, 15AR1747, 15AR0981 and T1–21 (pcjDM) were connected with gray
bars to demonstrate similar regions. We hypothesize the lagD and tetO genes were in the same region
and located in the plasmid of the ST-6964 ancestor strain, and the genome structure of the region in the
ancestral plasmid was similar to the equivalent region in the chromosome of isolate 15AR1747.
Subsequently, we hypothesize, this region was excised from the 15AR1747-m plasmid and integrated
into its chromosome; whereas in 15AR0984-m plasmid, only the tetO gene region was excised and
integrated into its chromosome, leaving lagD still in the 15AR0984-m plasmid. Alternatively, the tetO
could be carried by a pcJDM-like plasmid, and subsequently excised from the 15AR0981-m plasmid and
integrated the chromosomes (15AR0984 and 15AR1747 chromosomes).
